Random coefficient models
| Copyright notice |
|---|
| This article Linear mixed models (=Random Coefficient Models) was adapted from an original article by Nicholas Tibor Longford, which appeared in StatProb: The Encyclopedia Sponsored by Statistics and Probability Societies. The original article ([http://statprob.com/encyclopedia/RandomCoefficientModels.html StatProb Source], Local Files: pdf | tex) is copyrighted by the author(s), the article has been donated to Encyclopedia of Mathematics, and its further issues are under Creative Commons Attribution Share-Alike License'. All pages from StatProb are contained in the Category StatProb. |
2020 Mathematics Subject Classification: Primary: 62J10 [MSN][ZBL]
$ \newcommand{\refb}{\ref} $
$ \newcommand{\bbeta}{ { {\mathbf \beta}}} $ $ \newcommand{\bdelta}{ { {\mathbf \delta}}} $ $ \newcommand{\bomega}{ { {\mathbf \omega}}} $ $ \newcommand{\bOmega}{ { {\mathbf \Omega}}} $ $ \newcommand{\bSigma}{ { {\mathbf \Sigma}}} $ $ \newcommand{\tr}{\mbox{tr}} $ $ \newcommand{\tra}{^{\top}} $ $ \newcommand{\pri}{^{\prime}} $ $ \newcommand{\mE}{\mbox {E}} $ $ \newcommand{\bG}{ {\bf G}} $ $ \newcommand{\bH}{ {\bf H}} $ $ \newcommand{\bI}{ {\bf I}} $ $ \newcommand{\bUU}{ {\bf U}} $ $ \newcommand{\bV}{ {\bf V}} $ $ \newcommand{\bW}{ {\bf W}} $ $ \newcommand{\bX}{ {\bf X}} $ $ \newcommand{\bZ}{ {\bf Z}} $ $ \newcommand{\bY}{ {\bf Y}} $ $ \newcommand{\be}{ {\bf e}} $ $ \newcommand{\bi}{ {\bf i}} $ $ \newcommand{\bq}{ {\bf q}} $ $ \newcommand{\bs}{ {\bf s}} $ $ \newcommand{\buu}{ {\bf u}} $ $ \newcommand{\bx}{ {\bf x}} $ $ \newcommand{\by}{ {\bf y}} $ $ \newcommand{\bz}{ {\bf z}} $ $ \newcommand{\bzer}{ {\bf 0}} $ $ \newcommand{\var}{\mbox {var}} $ $ \newcommand{\cor}{\mbox {cor}} $
}
Summary
Random coefficient models are intended for settings with two or more sources of random variation. The widest range of applications is found for them when observational units form natural clusters, such that the units within a cluster are more similar than units in general. Models for independent observations have to be extended to allow for within- and between-cluster variation. \vspace*{1cm}
Keywords: Analysis of variance; borrowing strength; clusters;
correlation structure; empirical Bayes analysis;
longitudinal analysis; maximum likelihood; ordinary regression.
\clearpage
Independence of the observations is a key assumption of many standard statistical methods, such as analysis of variance (ANOVA) and ordinary regression, and some of its extensions. Common examples of data structures that do not fit into such a framework arise in longitudinal analysis, in which observations are made on subjects at subject-specific sequences of time points, and in studies that involve subjects (units) occurring naturally in clusters, such as individuals within families, school\-children within classrooms, employees within companies, and the like. The assumption of independence of the observational units is not tenable when observations within a cluster tend to be more similar than observations in general. Such similarity can be conveniently represented by a positive correlation (dependence).
This article describes an adaptation of the ordinary regression for clustered observations. Such observations require two indices, one for elements within clusters, $i=1, \ldots, n_{j\,}$, and another for clusters, $j=1, \ldots, m$. Thus, we have $n = n_1 + \cdots + n_m$ elementary units and $m$ clusters. The ordinary regression model $$ \label{ML1} y_{ij} \,=\, \bx_{ij\,}\bbeta + \varepsilon_{ij} \,, \tag{1}$$
with the usual assumptions of normality, independence and equal variance (homoscedasticity) of the deviations $\varepsilon_{ij\,}$, $\varepsilon_{ij} \sim {\cal{N}}(0, \sigma^2)$, i.i.d., implies that the regressions within the clusters $j$ have a common vector of coefficients $\bbeta$. This restriction can be relaxed by allowing the regressions to differ in their intercepts. A practical way of defining such a model is by the equation $$ \label{ML2} y_{ij} \,=\, \bx_{ij\,} \bbeta + \delta_j + \varepsilon_{ij} \,, \tag{2}$$
where $\delta_{j\,}$, $j=1, \ldots, m$, are a random sample from a centred normal distribution, $\delta_j \sim {\cal{N}}(0, \sigma^2_{\rm B})$, i.i.d., independent from the $\varepsilon$'s. This differs from the model for analysis of covariance (ANCOVA) only by the status of the deviations $\delta_{j\,}$. In ANCOVA, they are fixed (constant across hypothetical replications), whereas in \ref{ML2} they are random.
In the model in \ref{ML2}, the within-cluster regressions are parallel --- their intercepts are $\beta_0 + \delta_{j\,}$, but the coefficients on all the other variables in $\bx$ are common to the clusters. A more appealing interpretation of the model is that observations in a cluster are correlated, $$ \cor \left ( y_{i_1, j\,}, y_{i_2, j} \right ) \,=\, \frac{\sigma^2_{\rm B}} {\sigma^2 + \sigma^2_{\rm B}} \,, $$ because they share the same deviation $\delta_{j\,}$. Further relaxation of how the within-cluster regressions differ is attained by allowing some (or all) the regression slopes to be specific to the clusters. We select a set of variables in $\bx$, denoted by $\bz$, and assume that the regressions with respect to these variables differ across the clusters, but are constant with respect to the remaining variables; $$ \label{ML3} y_{ij} \,=\, \bx_{ij\,} \bbeta + \bz_{ij\,}\bdelta_j + \varepsilon_{ij} \,, \tag{3}$$
where $\bdelta_{j\,}$, $j=1, \ldots, m$, are a random sample from a multivariate normal distribution ${\cal{N}}(\bzer, \bSigma_{\rm B})$, independent from the $\varepsilon$'s. We say that the variables in $\bz$ are associated with (cluster-level) variation. The variance of an observation $y_{ij\,}$, without conditioning on the cluster $j$, is $$ \var \left ( y_{ij} \right ) \,=\, \sigma^2 + \bx_{ij} \bSigma_{\rm B\,} \bx_{ij} \tra \,. $$ We refer to $\sigma^2$ and $\bz_{ij} \bSigma_{\rm B\,} \bz_{ij}\tra$ as the variance components (at the elementary and cluster levels, respectively). The principle of invariance with respect to linear transformations of $\bz$ implies that the intercept should always be included in $\bz$, unless $\bz$ is empty, as in the model in \ref{ML1}. The function $V(\bz) = \bz \bSigma_{\rm B} \bz\tra$, over the feasible values of $\bz$, defines the pattern of variation, and it can be described by its behaviour (local minima, points of inflection, and the like). By way of an example, suppose $\bz$ contains the intercept and a single variable $z$. Denote the variances in $\bSigma_{\rm B}$ by $\sigma^2_0$ and $\sigma^2_{\rm z\,}$, and the covariance by $\sigma_{0\rm z\,}$. Then $$ \label{ML4} V(\bz) \,=\, \sigma^2_0 + 2z \sigma_{0\rm z} + z^2 \sigma^2_{\rm z} \,, \tag{4}$$
and this quadratic function has a unique minimum at $z^{\ast} = -\sigma_{0\rm z}/\sigma_{\rm z\,}^2$, unless $\sigma^2_{\rm z} =0$, in which case we revert to the model in \ref{ML2} in which $V(\bz)$ is constant. Figure 1 illustrates four patterns of variation on examples with a single covariate.
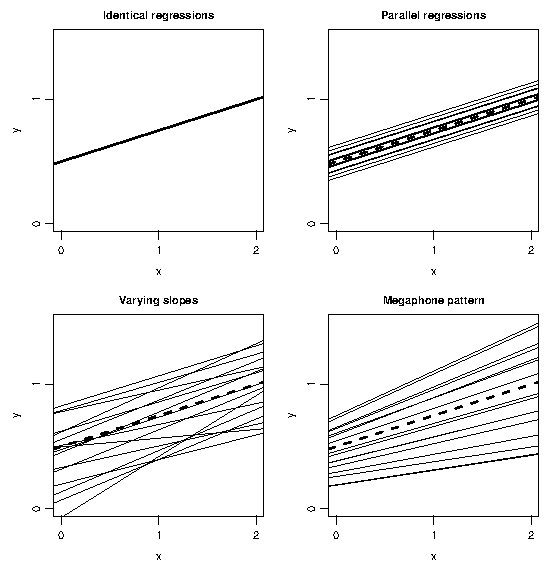
The model in \ref{ML3} is fitted by maximum likelihood (ML)
which maximizes the log-likelihood function1
$$ \label{Fisc1}
l \left (\bbeta, \sigma^2, \bSigma_{\rm B} \right ) \,=\,
-\frac{1}{2} \sum_{j=1}^m
\left [ \log \left \{ \det \left ( \bV_j \right ) \right \} \,+\,
\left ( \by_j - \bX_j \bbeta \right ) \tra \bV_j^{-1}
\left ( \by_j - \bX_j \bbeta \right ) \right ] \,,
\tag{5}$$
where $\bV_j$ is the variance matrix of the observations in cluster $j$, $\by_j$ the vector of the outcomes for the observations in cluster $j$, and $\bX_j$ the corresponding regression design matrix formed by vertical stacking of the rows $\bx_{ij\,}$, $i=1, \ldots, n_{j\,}$. The variation design matrices $\bZ_{j\,}$, $j = 1, \ldots, m$, are defined similarly; with them, $\bV_j = \sigma^2 \bI_{n_j} + \bZ_j \bSigma_{\rm B} \bZ_j \tra$, where $\bI_{n_j}$ is the $n_j \times n_j$ identity matrix. The Fisher scoring algorithm for maximising the log-likelihood in \ref{Fisc1} is described in the Appendix; for details and applications, see see Longford (1993), and for an alternative method Goldstein (2000). These and other algorithms are implemented in most standard statistical packages. A key to their effective implementation are closed-form expressions for the inverse and determinant of patterned matrices (Harville, 1997).
Model selection entails two tasks, selecting a set of variables to form $\bx$ and selecting its subset to form $\bz$. The variables in $\bx$ can be defined for elements or clusters; the latter can be defined as being constant within clusters. Inclusion of cluster-level variables in $\bz$ does not have an interpretation in terms of varying regression coefficients, so associating them with variation is in most contexts not meaningful. However, the identity in \ref{ML4} and its generalisations for $\bSigma_{\rm B}$ with more than two rows and columns indicate that $\bz$ can be used for modelling variance heterogeneity. The likelihood ratio test statistic and various information criteria can be used for selecting among alternative models, so long as one is a submodel of the other; that is, the variables in both $\bx$ and $\bz$ of one model are subsets of (or coincide with) their counterparts in the other model.
Random coefficients can be applied to a range of models much wider than ordinary regression. In principle, we can conceive any basis model, characterized by a vector of parameters, which applies to every cluster. A subset of these parameters is constant across the clusters and the remainder varies according to a model for cluster-level variation. The latter model need not be a multivariate normal distribution, although suitable alternatives to it are difficult to identify. The basis model itself can be complex, such as a random coefficient model itself. This gives rise to three- or, generally, multilevel models, in which elements are clustered within two-level units, these units in three-level units, and so on.
Generalized linear mixed models (GLMM) have generalized linear models (McCullagh and Nelder, 1989) as their basis; see Pinheiro and Bates (2000). For cluster $j$ we posit the model $$ g \left \{ \mE(\by_j\,|\,\bdelta_j) \right \} \,=\, \bX_j \bbeta + \bZ_j \bdelta_j \,, $$ with a (monotone) link function $g$, which transforms the expected outcomes to a linear scale; the outcomes (elements of $\by_j$) are conditionally independent given $\bdelta_j$ and have a specified distribution, such as binary or gamma. Some advantage, in both interpretation and computing, is gained by using canonical link functions, for which the conditional likelihood has a compact set of sufficient statistics. For example, the logistic link, $g(p) = \log(p)-\log(1-p)$, is the canonical link for binary outcomes, and the logarithm is the canonical link for Poisson outcomes. Models with the (standard) normality assumptions correspond to the link $g(y)=y$.
Without conditioning, the likelihood for non-normally distributed $\by_j$ has an intractable form. An established approach to fitting GLMM maximises a (tractable) normal-like approximation to the log-likelihood. This algorithm can be described as an iteratively reweighted version of the Fisher scoring (or another) algorithm for fitting the (normal) linear mixed model. With the advent of modern computing, using numerical quadrature and other methods for numerical integration has because feasible, especially for fitting models with univariate deviations $\delta_{j\,}$. An alternative framework for GLMM and a computational algorithm for fitting them are constructed by Lee and Nelder (2001).
Random coefficient models are well suited for analysing surveys in which clusters arise naturally as a consequence of the organisation (design) of the survey and the way the studied population is structured. They can be applied also in settings in which multiple observations are made on subjects, as in longitudinal studies (Molenberghs and Verbeke, 2000). In some settings it is contentious as to whether the clusters should be regarded as fixed or random. For example, small-area estimation (Rao, 2003) is concerned with inferences about districts or another partition of a country when some (or all) districts are represented in the analysed national survey by small subsamples. In one perspective, district-level quantities, such as their means of a variable, should be regarded as fixed because they are the inferential targets, fixed across hypothetical replications. When they are assumed to be random the (random coefficient) models are often more parsimonious than their fixed-effects (ANCOVA) counterparts, because the number of parameters involved does not depend on the number of clusters.
Borrowing strength (Robbins, 1955, Efron and Morris, 1972) is a general principle for efficient inference about each cluster (district) by exploiting the similarity of the clusters. It is the foundation of the empirical Bayes analysis, in which the between-cluster variance matrix plays a role similar to the Bayes prior for the within-cluster regression coefficients. The qualifier `empirical' refers to using a data-based estimator $\widehat{\bSigma}_{\rm B}$ in place of the unknown $\bSigma_{\rm B\,}$.
References
| [1] | Efron, B., and Morris, C. N. (1972). Limiting the risk of Bayes and empirical Bayes estimators --- Part II: empirical Bayes case. Journal of the American Statistical Association 67, 1286--1289. |
| [2] | Goldstein, H. (2000). Multilevel Statistial Models. 2nd Edition. London: Edward Arnold. |
| [3] | Harville, D. (1997). Matrix Algebra from a Statistician's Perspective. New York: Springer-Verlag. |
| [4] | Lee, Y., and Nelder, J. A. (2001). Hierarchical generalised linear models: a synthesis of generalised linear models, random effect models and structured dispersions. Biometrika 88, 987--1004. |
| [5] | Longford, N. T. (1993). Random Coefficient Models. Oxford: Oxford University Press. |
| [6] | Magnus, J. R., and Neudecker, H. (1988). Matrix Differential Calculus with Applications in Statistics and Econometrics. New York: Wiley. |
| [7] | McCullagh, P., and Nelder, J. A. (1989). Generalized Linear Models. 2nd Edition. London: Chapman and Hall. |
| [8] | Pinheiro, J. C., and Bates, D. M. (2000). Mixed-Effects Models in S and Splus. New York: Springer-Verlag. |
| [9] | Rao, J. N. K. (2003). Small Area Estimation. New York: Wiley and Sons. |
| [10] | Robbins, H. (1955). An empirical Bayes approach to statistics. Proceedings of the Third Berkeley Symposium on Mathematical Statistics and Probability, 1, 157--164. Berkeley, CA: University of California Press. |
| [11] | Verbeke, G., and Molenberghs, G. (2000). Linear Mixed Models for Longitudinal Data. New York: Springer-Verlag. |
- {1cm}
Appendix. Fisher scoring algorithm
This Appendix describes a method for fitting a random coefficient model by maximum likelihood. We prefer to use the scaled variance matrices $\bW_j = \sigma^{-2} \bV_j$ and $\bOmega = \sigma^{-2} \bSigma_{\rm B\,}$, so that $\bW_j = \bI_{n_d} + \bZ_j \bOmega \bZ_j \tra$ does not depend on $\sigma^2$. The log-likelihood in \ref{Fisc1} is equal to $$ \label{Fisc2} l \left (\bbeta, \sigma^2, \bOmega \right ) \,=\, -\frac{1}{2} \sum_{j=1}^m \left [ n \log \left ( \sigma^2 \right ) + \log \left \{ \det \left ( \bW_j \right ) \right \} \,+\, \frac{1}{\sigma^2} \, \be_j \tra \bW_j^{-1} \be_j \right ] \,. \tag{6}$$
where $\be_j = \by_j - \bX_j \bbeta$ is the vector of residuals for cluster $j$. We have the following closed-form expressions for the inverse and determinant of $\bW_{j\,}$: \begin{eqnarray} \label{FiscI} \bW_j^{-1} &=& \bI_{n_d} - \bZ_j \bOmega \bG_j^{-1} \bZ_j \tra \nonumber \\ \det \left ( \bW_j \right ) &=& \sigma^{2n_d} \det \left ( \bG_j \right ) \,, \end{eqnarray} where $\bG_j = \bI_{r} + \bZ_j \tra \bZ_j \bOmega$. All the matrices $\bG_j$ have the same dimension, $r \times r$, where $r$ is the number of variables in $\bZ$.
Assuming that the log-likelihood $l$ has a single maximum, it can be found as the root of the score vector. By matrix differentiation we obtain $$ \frac{\partial l}{\partial \bbeta} \,=\, \frac{1}{\sigma^2} \sum_{j=1}^m \bX_j \tra \bW_j^{-1} \be_j \,, $$ and so the maximum likelihood estimator of $\bbeta$ is the generalised least-squares estimator $$ \label{FiscB} \hat{\bbeta} \,=\, \left ( \sum_{j=1}^m \bX_j \tra \hat{\bW}_j^{-1} \bX_j \right )^{-1} \sum_{j=1}^m \bX_j \tra \hat{\bW}_j^{-1} \by_j \,. \tag{7}$$
The matrices $\hat{\bW}_j$ are the estimated versions of $\bW_{j\,}$, with estimator $\hat{\bOmega}$ substituted for $\bOmega$. Estimation of $\bOmega$ is described below.
The elementary-level (residual) variance $\sigma^2$ is estimated by the root of its score, $$ \frac{\partial l}{\partial \sigma^2} \,=\, -\frac{1}{2} \left ( \frac{n}{\sigma^2} - \frac{1}{\sigma^4} \sum_{j=1}^m \be_j \tra \bW_j^{-1} \be_j \right ) \,, $$ which is $$ \hat{\sigma}^2 \,=\, \frac{1}{n} \sum_{j=1}^m \be_j \tra \hat{\bW}_j^{-1} \be_j \,. $$ The elements of $\bOmega$ are estimated by the Fisher scoring algorithm. Let these elements, without any redundancy, comprise the vector $\bomega$. In most applications, $\bomega$ comprises the $\frac{1}{2} r \times (r+1)$ unique elements, $r$ variances and $\frac{1}{2} r \times (r-1)$ covariances. The Fisher scoring algorithm proceeds by iterations that update the estimate of $\omega$, based on the score vector $\bs$ and the expected information matrix $\bH$ evaluated at the current solution. The vector $\bs$ and matrix $\bH$ are derived by matrix differentiation. See Magnus and Neudecker (1988) and Harville (1997) for background.
Let $\bi_h$ be the ($r \times 1$) indicator vector for element $h$. For example, when $r = 5$, $\bi_2 = (0,1,0,0,0)\tra$. The element of $\bomega$ that corresponds to the (scaled) covariance $(h, h\pri)$ in $\bOmega$ can be expressed as $$ \omega \,=\, \bi_h \tra \bOmega \bi_{h\pri} \,, $$ and $\partial \bOmega /\partial \omega = \bi_{h\,}\bi_{h\pri}\tra + \bi_{h\pri\,}\bi_{h}\tra$. If we replace each (scaled) variance in $\bomega$ by its half, then this expression holds also for these half-variance parameters. With this parametrisation, noting that $\partial \bW_j /\partial \omega = \bZ_{j\,} \partial \bOmega/ \partial \omega_{\,} \bZ_j\tra$, we have \begin{eqnarray*} \frac{\partial l}{\partial \omega} &=& - \frac{1}{2} \sum_{j=1}^m \left \{ \tr \left ( \bW_j^{-1} \frac{\partial{\bW_j}}{\partial \omega} \right ) - \frac{1}{\sigma^2} \, \be_j \tra \bW_j^{-1} \frac{\partial{\bW_j}}{\partial \omega} \bW_j^{-1} \be_j \right \} \\ &=& \sum_{j=1}^m \left ( - \bi_h \tra bUU_{j\,} \bi_{h\pri} + \frac{1}{\sigma^2}\, \buu_j \tra \bi_h \, \buu_j \tra \bi_{h\pri} \right ) \\ &=& \sum_{j=1}^m \left ( - U_{j,hh\pri} + \frac{1}{\sigma^2}\, u_{j,h\,}u_{j,h\pri} \right ) \,, \end{eqnarray*} where $bUU_j = \bZ_j \tra \bW_j^{-1} \bZ_{j\,}$, $\buu_j = \bZ_j \tra \bW_j^{-1} \be_{j\,}$, $U_{j,hh\pri}$ is the $(h,h\pri)$-element of $bUU_j$ and $u_{j,h}$ the $h$-element of $\buu_{j\,}$. Multiplying by a vector $\bi_h$ amounts to extracting an element. Thus, evaluation of the score $\partial l/\partial \omega$ requires calculation of quadratic forms $\bq \tra \bW_j\tra\bZ_{j\,}$. From \ref{FiscI} we have, for an arbitrary $n_d \times 1$ vector $\bq$, the identity $$ \bZ_j \tra \bW_j^{-1} \bq \,=\, \bG_j ^{-1} \bZ_j \tra \bq \,, $$ so we do not have to form the matrices $\bW_j$ and do not have to invert any matrices of large size. Further differentiation yields the expression { \begin{eqnarray*} \frac{\partial ^2 l}{\partial \omega_{1\,} \partial \omega_2} &=& \sum_{j=1}^m \left [ \bi_{h_1} \tra bUU_{j\,} \frac{\partial \bOmega}{\partial \omega_2} \, bUU_j \bi_{h\pri_1} \,-\, \frac{2}{\sigma^2} \left \{ \buu_j \tra \bi_{h_1\,} \buu_j \tra \frac{\partial \bOmega}{\partial \omega_2} \, bUU_j \bi_{h\pri_1} + \buu_j \tra \bi_{h\pri_1\,} \buu_j \tra \frac{\partial \bOmega}{\partial \omega_2} \, bUU_j \bi_{h_1} \right \} \right ] \\ &=& \sum_{j=1}^m \left \{ U_{j,h_1h_2\,}U_{j,h\pri_1h\pri_2\,} + U_{j,h\pri_1h_2\,}U_{j,h_1h\pri_2\,} \right. \\ && - \left. \! \frac{2}{\sigma^2}\, \left ( u_{j,h_1\,}u_{j,h_2\,} U_{j,h\pri_1h\pri_2} + u_{j,h_1\,}u_{j,h\pri_2\,} U_{j,h\pri_1h_2} + u_{j,h\pri_1\,}u_{j,h_2\,} U_{j,h_1h\pri_2} + u_{j,h\pri_1\,}u_{j,h\pri_2\,} U_{j,h_1h_2} \right ) \right \} \end{eqnarray*}} \vspace*{-6mm}
for $\omega_1$ and $\omega_2$ associated with the respective
elements $(h_{1\,}, h\pri_1)$ and $(h_{2\,}, h\pri_2)$ of $\bOmega$.
The Hessian matrix, its negative expectation, comprises elements
\begin{eqnarray*}
H \left ( \omega_{1\,},\omega_2 \right ) &=& \sum_{j=1}^m \left (
\bi_{h_1} \tra bUU_j \bi_{h_2} \,
\bi_{h\pri_1} \tra bUU_j \bi_{h\pri_2} \,+\,
\bi_{h\pri_1} \tra bUU_j \bi_{h_2} \,
\bi_{h_1} \tra bUU_j \bi_{h\pri_2} \right )
\\ &=&
\sum_{j=1}^m \left ( U_{j,h_1h_2\,}U_{j,h\pri_1h\pri_2} +
U_{j,h\pri_1h_2\,}U_{j,h_1h\pri_2} \right ) \,,
\end{eqnarray*}
which are cluster-level totals of the cross-products of various
elements of $bUU_{j\,}$.
In each iteration $t$, the estimate of $\bomega$ is updated as
$$
\hat{\bomega}^{(t)} \,=\, \hat{\bomega}^{(t-1)} +
r \hat{\bH}_t^{-1} \hat{\bs}_t \,,
$$
where $r=1$, unless the updated matrix $\bOmega^{(t)}$ is not
positive definite.
One way to avoid this is to keep halving $r$ until the updated
matrix $\bOmega^{(t)}$ is positive definite. Having to do so
in many (or all) iterations is usually a sign of having included
too many variables in $\bZ$, and the model should be revised
accordingly.
An alternative approach estimates the Cholesky (or another)
decomposition of $\bOmega$. The iterations are terminated when
the norm of the updating vector $\bH^{-1}\bs$ is sufficiently small.
\vspace*{10mm}
Based on an article from Lovric, Miodrag (2011),
International Encyclopedia of Statistical Science.
Heidelberg: Springer Science + Business Media, LLC.
Linear mixed models. Encyclopedia of Mathematics. URL: http://encyclopediaofmath.org/index.php?title=Linear_mixed_models&oldid=37921